Post-transcriptional modification - RNA interference (RNAi)/ Alternative Splicing
Laura Armstrong & Joe Wolfensohn
Teachers


Contents
Recall Questions
This topic requires prior knowledge of transcription and translation from year 12. You can test your knowledge on these below.
What organelle is the site of translation?
The ribosome.
What does tRNA do during translation?
tRNA carries specific amino acids to the ribosome and the anticodon binds to a complementary codon on mRNA.
What determines the sequence of amino acids in a protein?
The sequence of codons in the mRNA / the sequences of bases in DNA
Topic Explainer Video
Check out this @JoeDoesBiology video that explains post-transcriptional modification - RNA interference (RNAi)/ alternative splicing or read the full notes below. Once you've gone through the whole note, try out the practice questions!
Introduction to Post-transcriptional modification
After a gene is transcribed into pre-mRNA, it does not always lead directly to protein production. Instead, gene expression can be regulated after transcription but before translation. This is known as post-transcriptional control, and it includes:
- RNA interference (RNAi) – involving siRNA
- Alternative splicing of pre-mRNA
RNA Interference (RNAi) via siRNA
What is siRNA?
- siRNA stands for small interfering RNA.
- It is a short, double-stranded RNA molecule (~20-25 base pairs long) that regulates gene expression after transcription, by preventing translation.
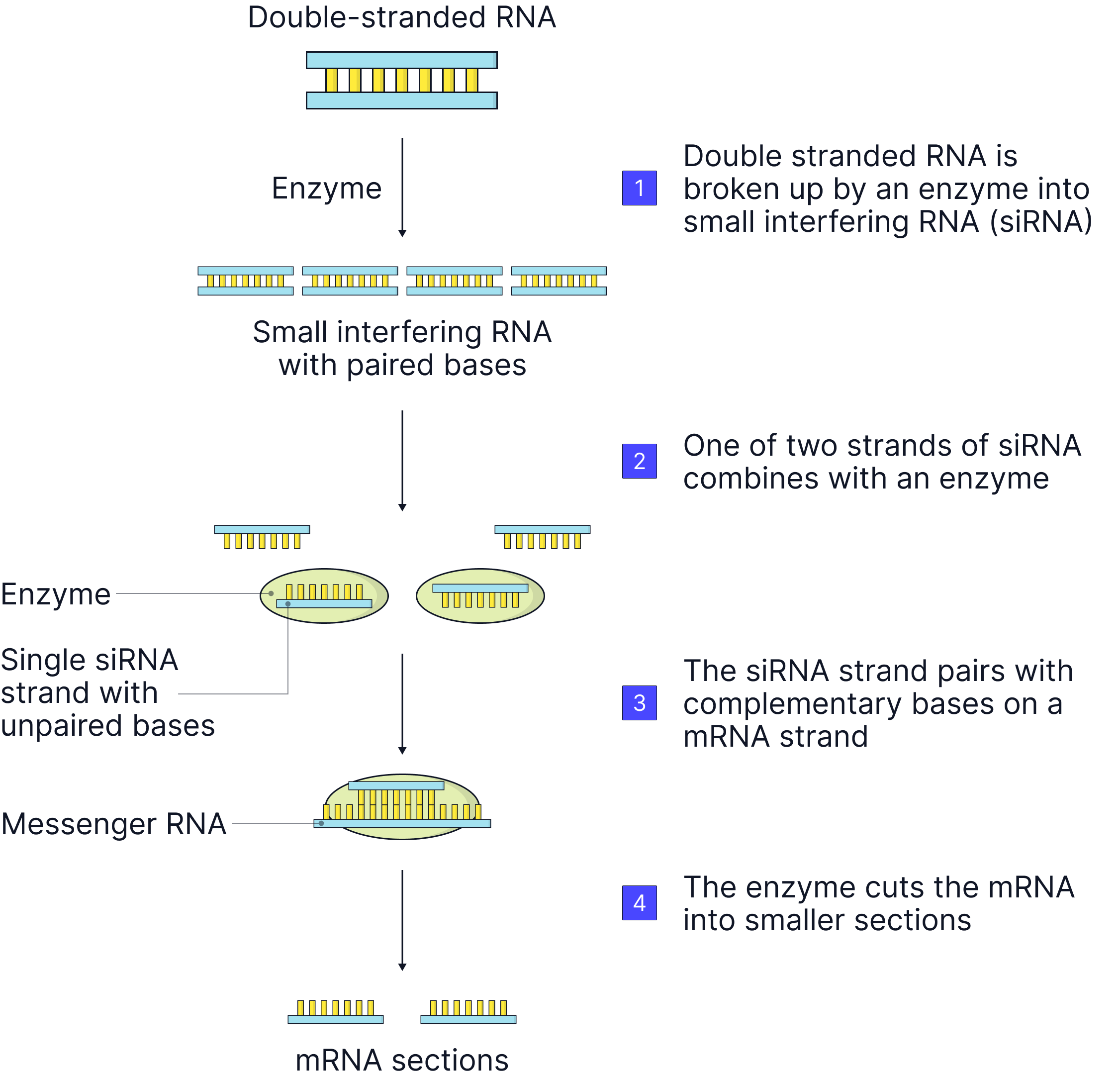
How siRNA Works:
- siRNA is produced from double-stranded RNA in the cell or introduced synthetically.
- It binds to a protein complex called the RNA-induced silencing complex.
- The siRNA is separated into single strands, and the one strand remains attached to the protein complex.
- This strand is complementary to a specific mRNA molecule.
- siRNA guides the protein complex to the mRNA and binds to it via complementary base pairing.
- An enzyme in the protein complex cuts the mRNA, breaking it into pieces.
- The mRNA is degraded, so translation cannot occur, and the gene is effectively silenced.
Result: The gene is transcribed, but it cannot be translated, therefore the protein cannot be produced.
Alternative Splicing
What Is It?
- After transcription, eukaryotic pre-mRNA contains both exons (coding regions) and introns (non-coding regions).
- Splicing removes introns and joins exons together to make mature mRNA.
Alternative Splicing:
- Different combinations of exons are joined together in different cells or conditions.
- This produces multiple forms of mRNA from a single gene.
- Each mRNA codes for a different protein with potentially different functions.
Result: A single gene can give rise to multiple proteins depending on how it’s spliced.
Why Is This Important?
| Mechnism | Control Point | Outcome |
|---|---|---|
|
siRNA |
After transcription |
mRNA is degraded → gene expression switched off (no protein produced) |
|
Alternative Splicing |
Between transcription and translation |
Different mRNA / proteins from one gene |
Key Terms
- siRNA: Small interfering RNA that binds to mRNA and causes its degradation.
- RNA interference (RNAi): A process where small RNA molecules inhibit gene expression by destroying mRNA.
- Alternative Splicing: Process by which different combinations of exons are joined to produce different mRNAs.
- Pre-mRNA: The initial RNA transcript before introns are removed by splicing.
- Post-transcriptional control: Regulation of gene expression after transcription but before translation.
Exam Tip
When answering questions on siRNA:
- Be precise with terms like binds, complementary base pairing, mRNA degradation, and inhibits translation.
- Don’t confuse siRNA with transcription factors — siRNA acts after transcription.
Scientists made siRNA to inhibit expression of a specific HIV gene inside a human cell. They attached this siRNA to a carrier molecule.
The flow chart shows what happens when this carrier molecule reaches a human cell infected with HIV.

This siRNA would only affect gene expression in cells infected with HIV.
Suggest two reasons why (4 marks)
- Only infected cells have HIV protein on surface
- So carrier only attaches to these cells / siRNA can only enter these cells
- siRNA base sequence complementary / specific to this mRNA
- Only infected cells contain mRNA of HIV / this gene
Practice Question
Try to answer the practice question from the TikTok on your own, then watch the video to see how well you did!